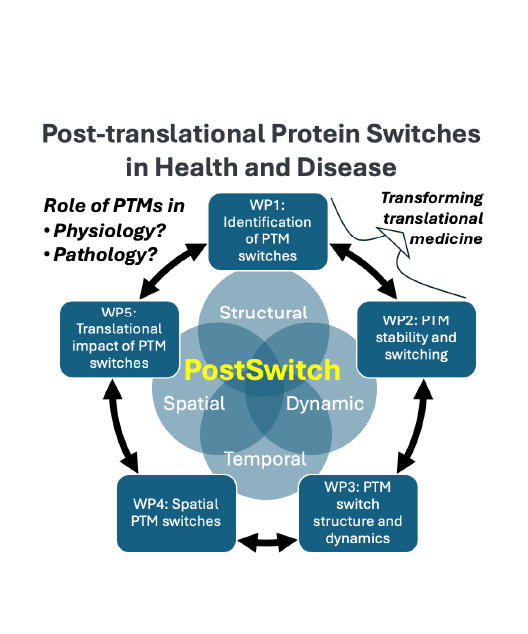
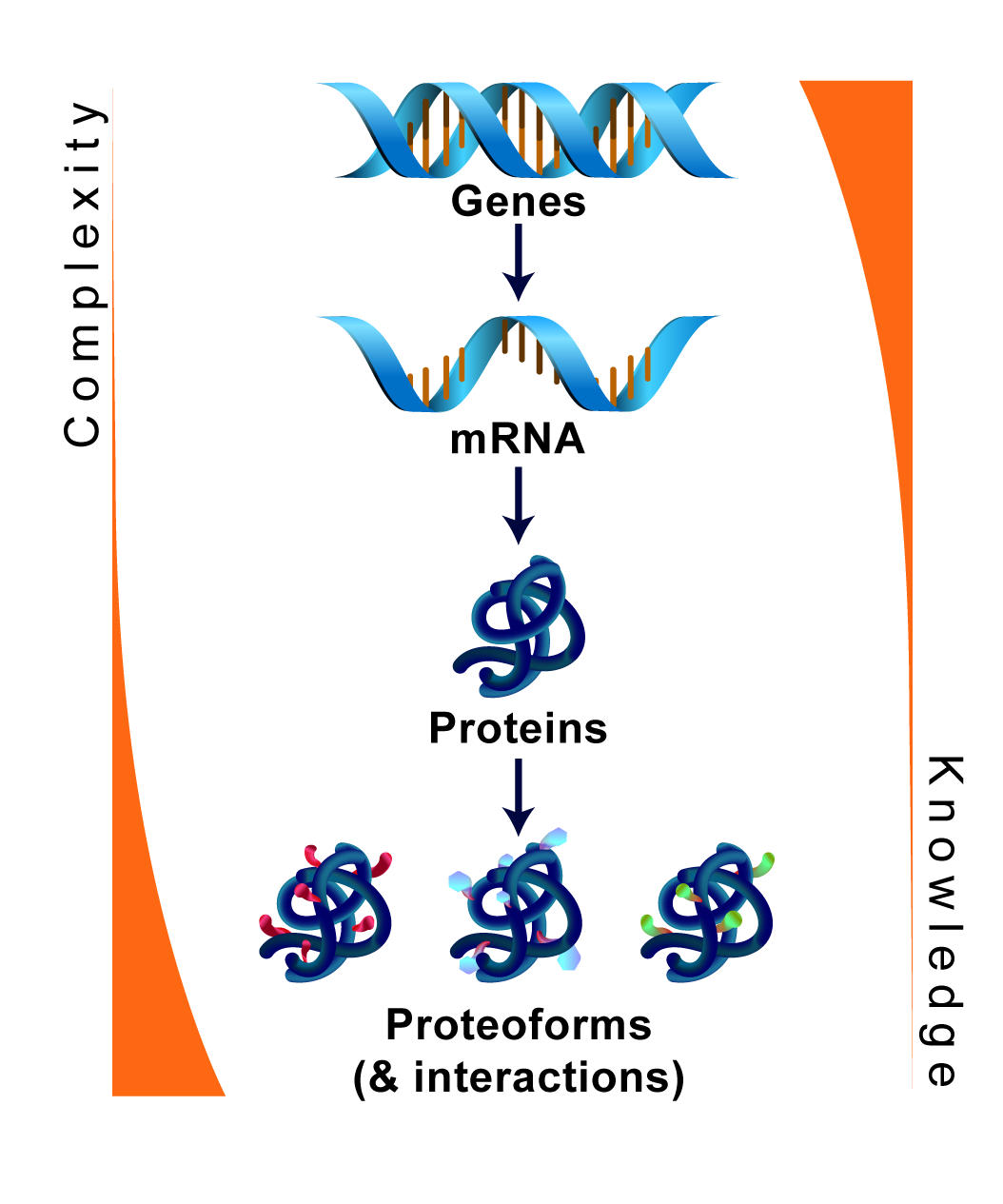
PostSwitch
All physiological and pathological conditions are governed by post-translationally modified (PTM) proteins that exert their functions in the context of protein complexes. PTMs have developed during evolution to meet the increasingly complex demands for fine tuning of the cellular functions in response signals the cells receive, but also to defend against impulses that might be deleterious to our cells. Switch-like PTM regulation provides thus a temporally and spatially tunable mode of regulation of cellular functions, but this cannot be readily deciphered by studying genomics. Centre of Excellence in post-translational protein switches in health and disease (PostSwitch) bridges critical gaps in our current understanding post-translational protein regulation via cross-disciplinary approaches. Join us on this journey to revolutionize our understanding of biology at the molecular level.
Unveiling the Mysteries of Protein Modifications:
Our research focuses on the fascinating journey of post-translational modification of proteins. By leveraging state-of-the-art proteomics, structral biology, spatial biology and computational modelling, we aim to unravel the complexities of protein modifications and their roles in cellular processes and diseases. As PTM modified proteins are the actual effectors in diseases, we believe that our findings will pave the way for groundbreaking advancements in medical and biotechnological fields.
Principal investigators
Our project is spearheaded by a team of distinguished leaders with extensive expertise in proteomics and protein research. With a rich background in both clinical and translational research, our leaders have made significant contributions in academia and industry. Their combined experience and innovative approaches drive our mission to decode the complexities of protein formation and its implications for health and disease.
Pioneering Insights in Proteomics and Protein Research
The leaders of our project have made significant scientific contributions through their extensive publications in top-tier journals. Their work has not only enhanced our understanding of fundamental biological processes but also paved the way for innovative therapeutic approaches to treat various diseases. Their publications are widely cited, reflecting the impact and importance of their research in both academia and industry.
Key publications
- Chemotherapy induces myeloid-driven spatially confined T cell exhaustion in ovarian cancer. Cancer Cell, 2024
- Structural basis of respiratory complex adaptation to cold temperatures. Cell, 2024
- Structural basis of mitochondrial membrane bending by the I–II–III2–IV2 supercomplex. Nature, 2023
- Horizontal proton transfer across the antiporter-like subunits in mitochondrial respiratory complex I. Chemical Science, 2023
- Structural mechanism for inhibition of PP2A-B56α and oncogenicity by CIP2A. Nature Communications, 2023
- A competitive precision CRISPR method to identify the fitness effects of transcription factor binding sites. Nature Biotechnology, 2023
- Single-cell tumor-immune microenvironment of BRCA1/2 mutated high-grade serous ovarian cancer. Nature Communications, 2022
- Optimized detection of somatic allelic imbalances specific for homologous recombination deficiency improves the prediction of clinical outcomes in cancer. NPJ Precision Oncology, 2022
- Human transcription factor protein interaction networks. Nature Communications, 2022.
- Physical and functional interactome atlas of human receptor tyrosine kinases. EMBO Reports, 2022
- The PP2A-Integrator-CDK9 axis fine-tunes transcription and can be targeted therapeutically in cancer. Cell, 2021
- The hantavirus surface glycoprotein lattice and its fusion control mechanism. Cell, 2020
- Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus. Nature, 2019
- PP2A inhibition is a druggable MEK inhibitor resistance mechanism in KRAS-mutant lung cancer cells. Science Translational Medicine, 2018




%20-%20THL.png)